My research is centered on a provocative question: how predictable is evolution? For if we can more accurately predict evolutionary trajectories, we might be better equipped to direct evolution down paths to desirable outcomes such as increased drug susceptibility: a worthwhile goal that can lead to improvements in disease treatment and quality of care. However, to predict the evolution of resistance with accuracy we must understand and integrate information about many factors, including a population’s evolutionary history, size, and mutation rate.
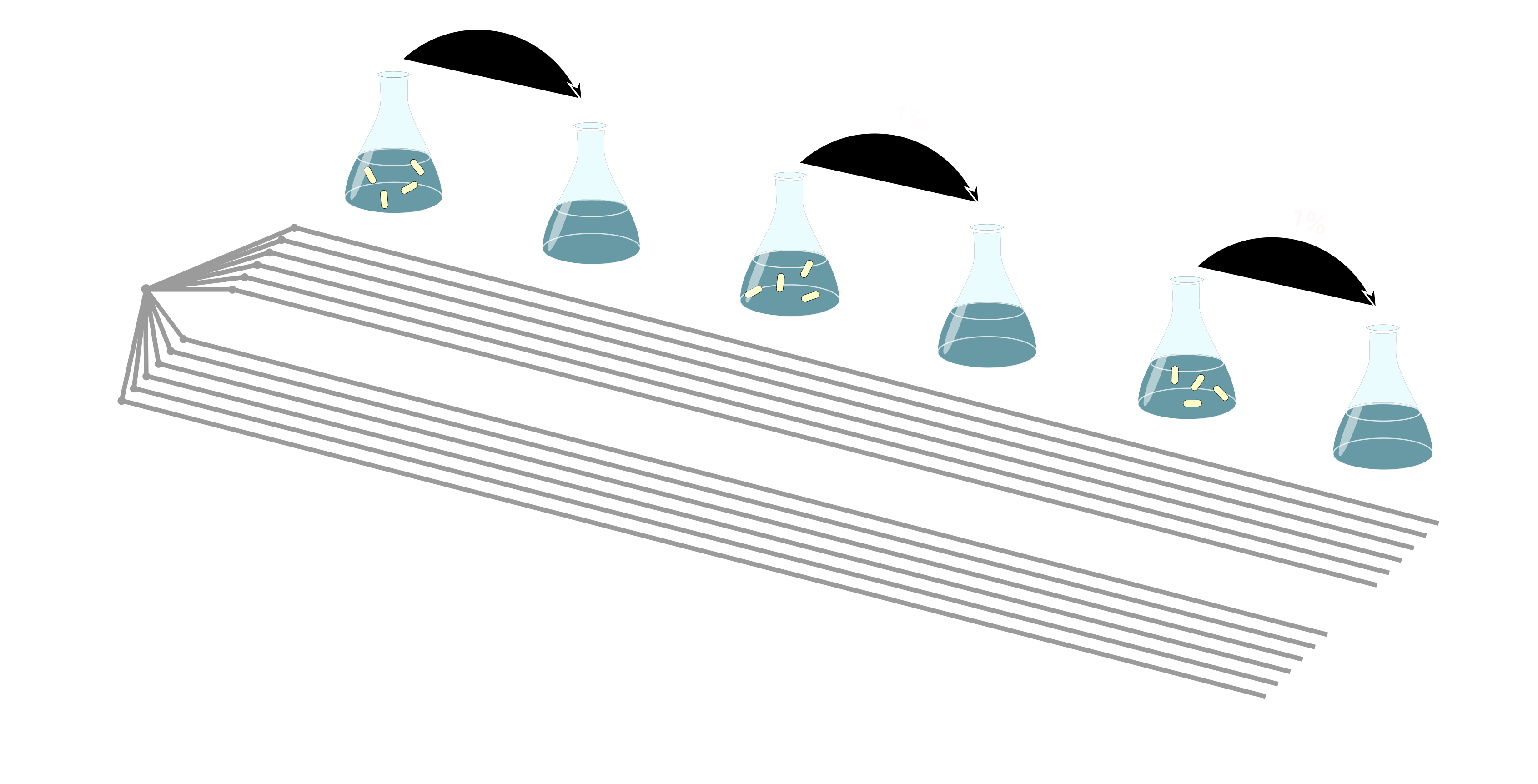
During my dissertation studies, I addressed how genetic background affects the evolution of phenotypic antibiotic resistance, its genetic basis, and its fitness costs using Escherichia coli strains from the long-term evolution experiment (LTEE) that independently evolved for multiple decades in an environment without antibiotics.
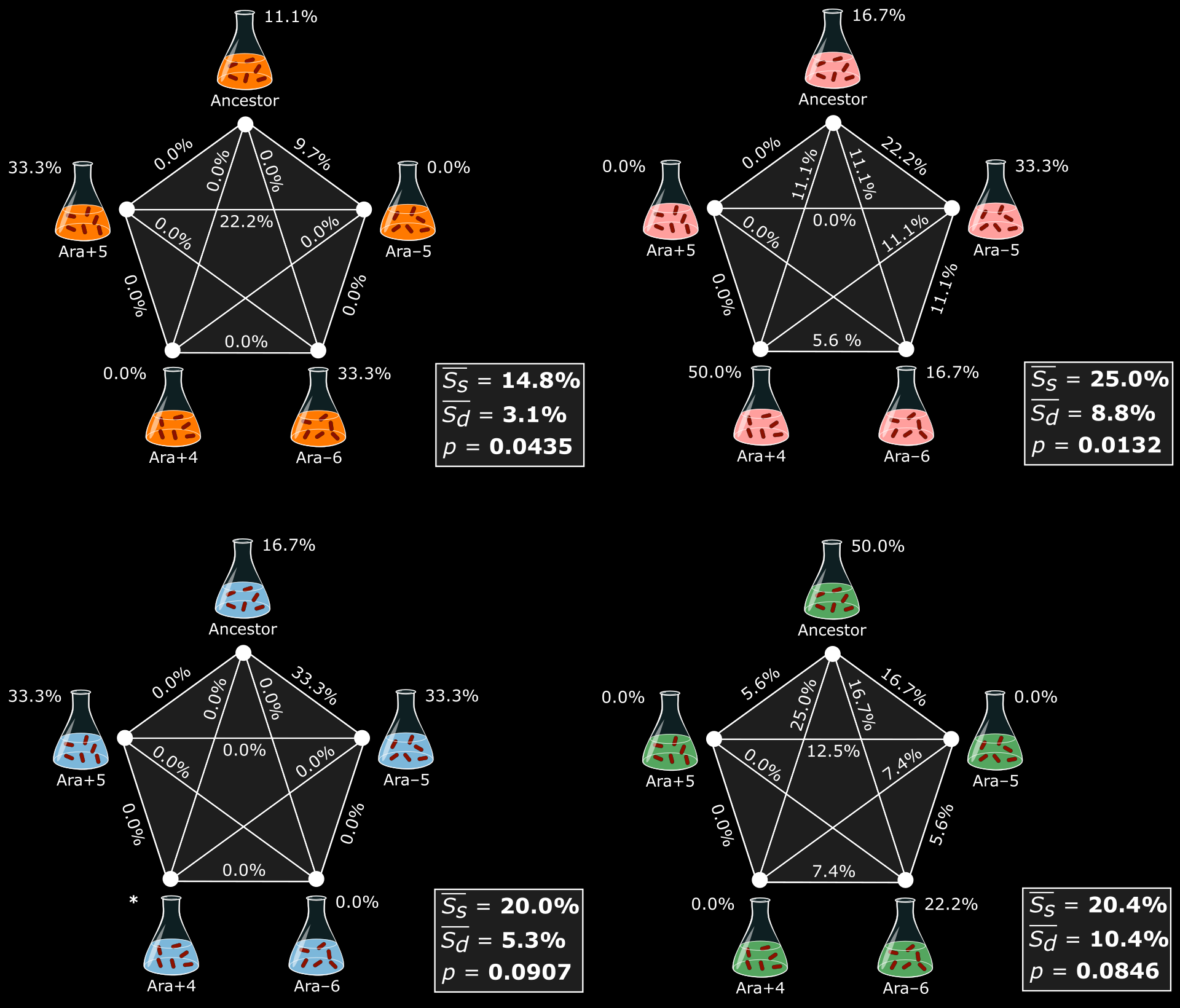
First, I examined how readily these LTEE strains could overcome prior losses of intrinsic resistance through subsequent evolution when challenged with antibiotics. Evolvability was idiosyncratic with respect to initial genotype, such that resistance was more constrained in some backgrounds than in others. Second, I investigated whether lineages founded from different genotypes take parallel or divergent mutational paths to achieve increased resistance. On average, replicate lines evolved from the same founding genotypes had more mutations in common at the gene level than did lines evolved from different founding genotypes. Third, I tested whether fitness costs of resistance mutations are constant across different genetic backgrounds. Resistant lines had an overall reduction in fitness relative to their sensitive counterparts in the absence of the drug, and there was significant among-line heterogeneity in fitness costs. These idiosyncratic differences were explained by mutations in different genetic targets, mutations in different genes that comprise the same physiological pathway, and even different alleles of the same gene. Taken together, my dissertation studies demonstrate that a lineage’s evolutionary history can alter both its phenotypic and genotypic paths to antibiotic resistance, and the associated fitness costs of resistance mutations.
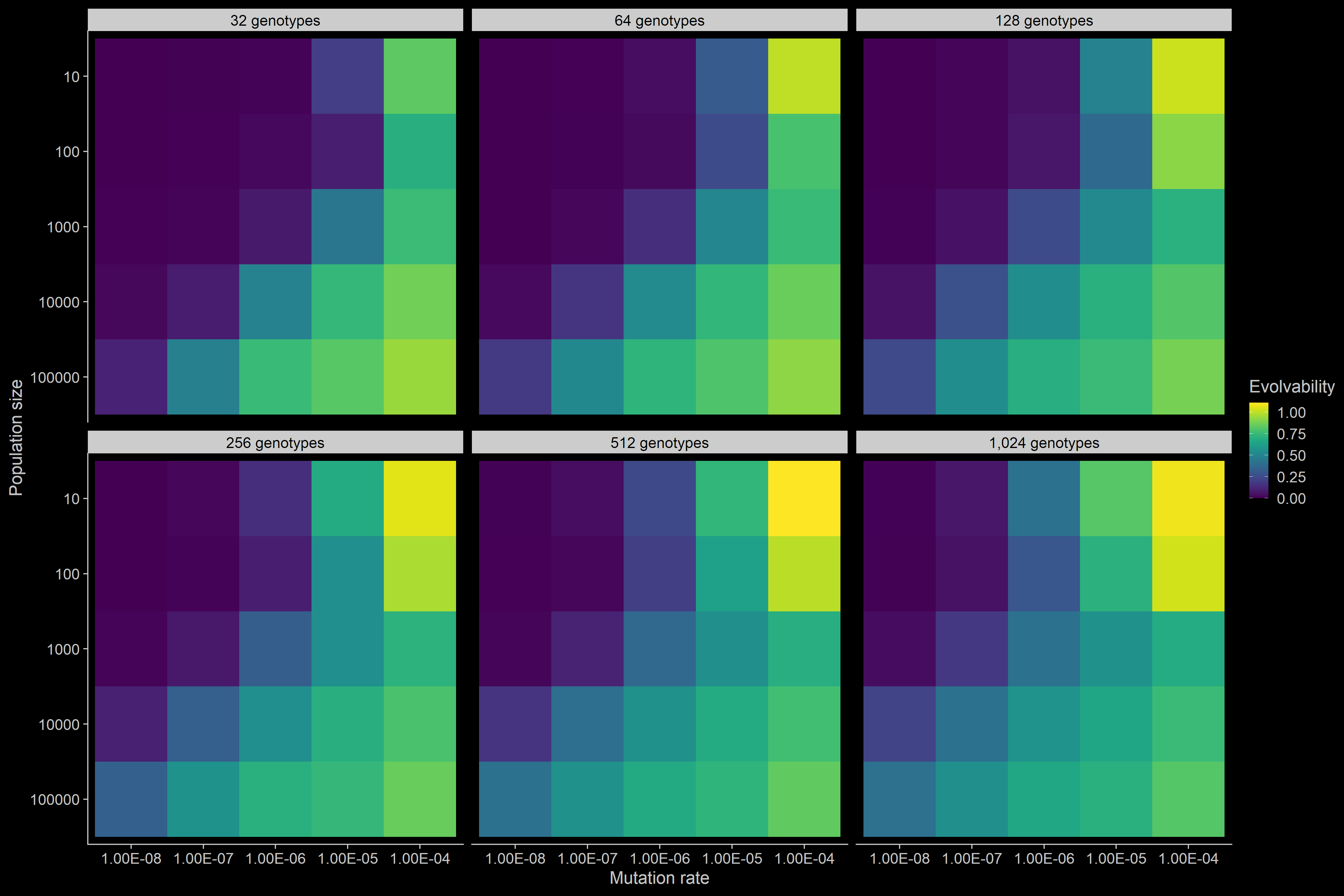
Nonetheless, it is still not clear how background integrates with demographic and other factors to influence resistance evolution. For example, rare pathways leading to high-level resistance may become more accessible with larger population sizes and increased mutation rates, but can this increase in evolutionary potential outweigh, or be outweighed by, any potentiating or constraining effects of changes in genetic background? I am currently addressing these issues as a postdoctoral researcher by using both stochastic simulations and bacterial and cancer model systems. Moreover, I am examining whether resistance potential is conserved across drug environments that are more similar (i.e., within-class drugs) relative to environments that are less similar (i.e., between-class drugs). This work underscores the need to consider the multifactorial nature of resistance evolution to predict, and ultimately control, this phenotype.