Predict. Control. Cure.
Exploring the genetic and ecological factors driving antibiotic resistance and pioneering new technologies to design effective, evolution-inspired therapeutic strategies.
I am dedicated to understanding and combating antibiotic resistance. My research combines experimental evolution, functional genomics, and computational modeling to identify the genetic and environmental factors that drive resistance. By integrating these approaches, I aim to develop innovative strategies to predict and guide the evolutionary trajectories of pathogens, ultimately improving treatment outcomes.
I am a disabled scientist born with a rare neurological condition called Moebius syndrome, which affects the muscles controlling facial expressions and eye movement. I also have several limb differences. Although my disabilities pose challenges, my identity as a disabled person has enriched my academic life in many ways. Through my lived experiences, I appreciate that, despite our differences, our curiosity about the natural world binds us together, each of us has a unique story that should be respected, and we all deserve a voice in science. I honor these ideals by giving my time to advocacy, mentorship, outreach, and learning.
I am committed to creating and maintaining an environment in which all are welcome and respected—one that is inclusive of race, gender, faith, sexual orientation, ability, and socioeconomic status.
Kyle Card, Ph.D.
HHMI Hanna H. Gray Postdoctoral Fellow

Evolution, History, and Control in Antibiotic Resistance
Connecting discoveries to a clinic-ready vision
Limiting resistance in the clinic.
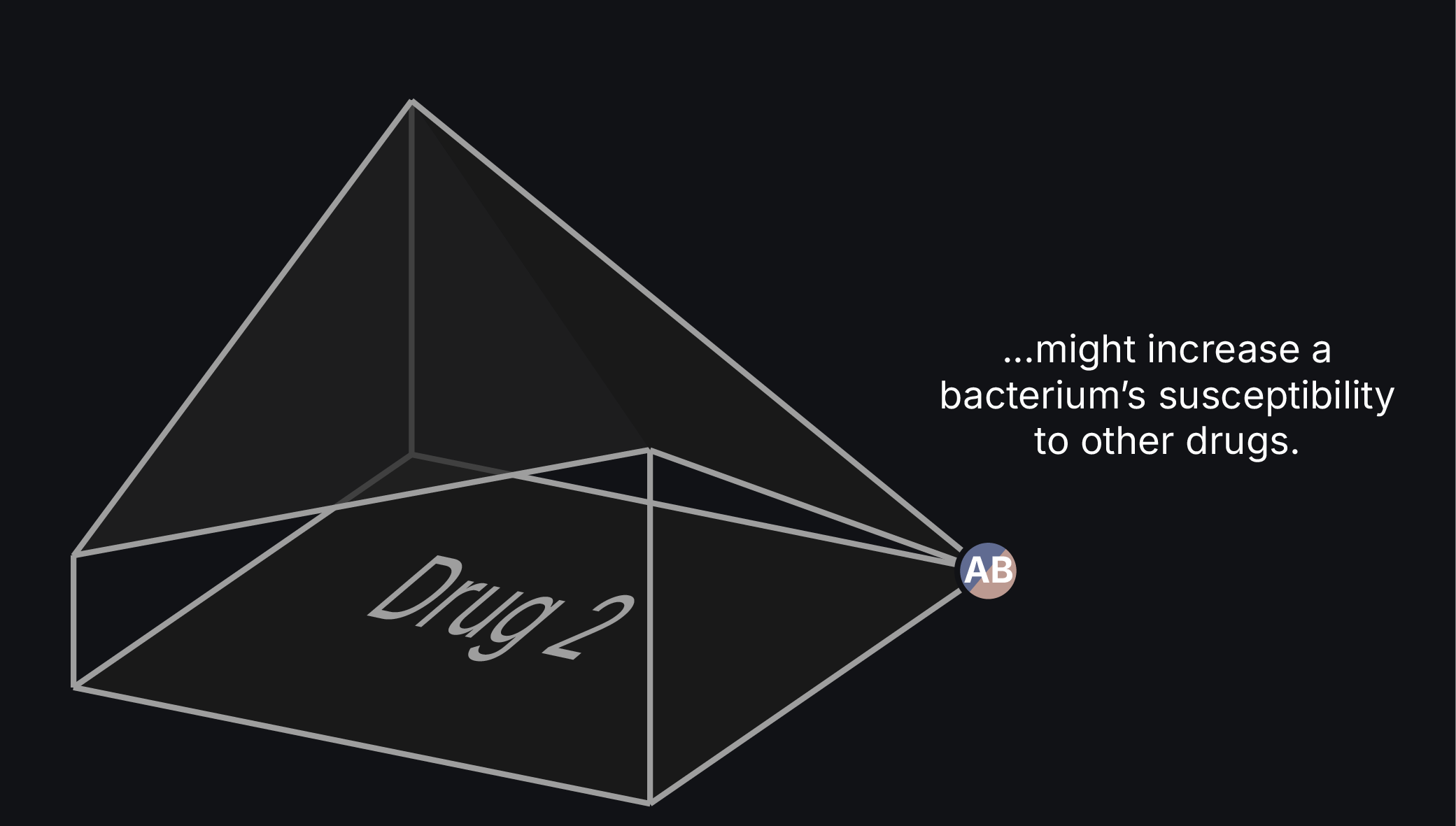
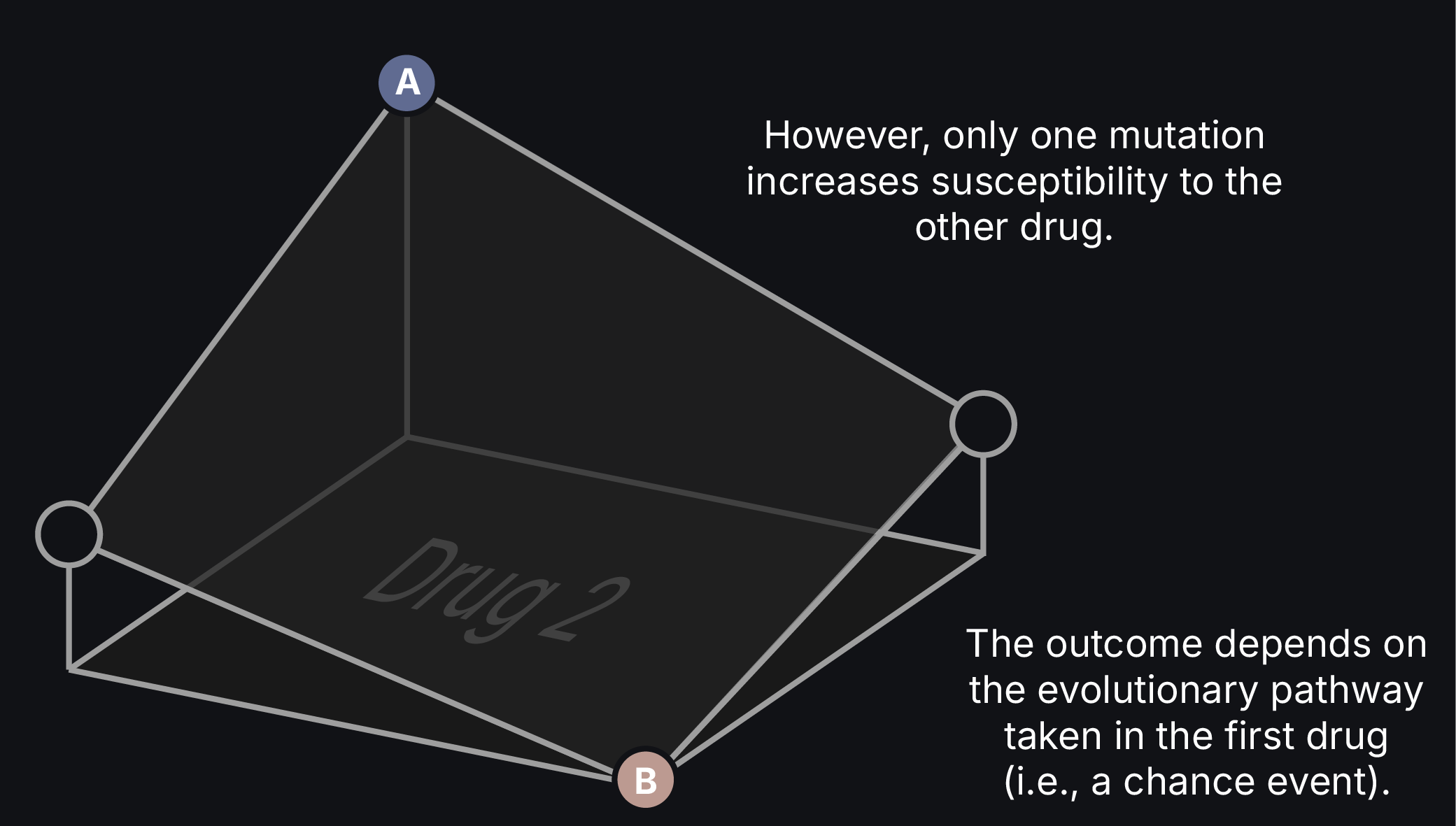
Imagine a scenario where a clinician strategically uses existing antibiotics to prevent or reverse drug resistance in an infection. This approach should be possible by exploiting drug tradeoffs in which the evolution of resistance to one therapy increases susceptibility to others. They would only need to alternate to one of these other drugs at the appropriate time.
However, interactions between mutations and their genetic backgrounds can open new adaptive routes or restrict others, challenging our ability to predict in which direction a population will evolve. We must, therefore, comprehensively understand how these interactions shape resistance evolution to design effective treatment strategies.
History matters.
As an HHMI Gilliam Fellow, I investigated how a bacterium's prior history influences its ability to evolve antibiotic resistance. Using E. coli strains from the Long-Term Evolution Experiment, we found that genomic differences between lines can unpredictably alter resistance and channel evolution down particular mutational pathways.
Although this work implicates genetic interactions in our ability to forecast resistance evolution, it is insufficient to focus solely on model organisms. We must systematically investigate these relationships in pathogens, bridging the divide between bench and bedside.
From model systems to pathogens.
As an HHMI Hanna H. Gray Fellow, I explored drug tradeoffs in S. aureus. We discovered that this opportunistic pathogen followed at least two distinct adaptive pathways as it evolved vancomycin resistance. These separate paths led to contrasting, yet predictable sensitivities to second-line antibiotics. To account for this uncertainty, we developed a framework called the Collateral Response Score (CRS) to provide a probabilistic forecast of how past antibiotic exposure might affect future treatment outcomes.
THE CARD LAB:
Where persistence meets resistance.
We established that a pathogen's evolutionary history is key to forecasting its adaptability and therapeutic responses. However, evolution is context-dependent. The selective pressures inside a flask are vastly different from those inside the complex, dynamic environment of the human body, where evolution occurs. To develop effective, evolution-informed therapies, we must bridge the gap between reductionist lab experiments and the clinical reality of infections.
My laboratory will pioneer the use of organ-chip technology to overcome this challenge. By integrating experimental evolution in these high-fidelity systems with functional genomics and phenotypic screens, we will dissect the molecular and evolutionary mechanisms of pathogen adaptation in real-time, creating a framework to predict and ultimately steer the evolution of antimicrobial resistance. We will focus on chronic P. aeruginosa infections in cystic fibrosis, an area with an unmet clinical need and an ideal model system for studying within-host evolution.
The Team
-

Justin Creary
GRADUATE STUDENT, CWRU
Justin is investigating how prior evolutionary history under varying concentrations of tobramycin affects the ability of P. aeruginosa to evolve resistance to clinically relevant second-line drugs.
-

Amira Stocks
UNDERGRADUATE STUDENT, CWRU
Amira is investigating the impact of biofilm formation and population bottleneck size on the evolutionary trajectories of antibiotic resistance in P. aeruginosa and how these pathways differ from planktonic populations.
-

Shiva Ayyar
UNDERGRADUATE STUDENT, CWRU
Shiva is examining the combined impact of genetic background and mutation supply on the potential of P. aeruginosa to evolve resistance to several first-line antibiotics.
-

Rohan Desai
UNDERGRADUATE STUDENT, VANDERBILT
Rohan joined our lab this summer and is working with Justin and Shiva to knock out the methyl-directed mismatch repair gene mutS in P. aeruginosa, thereby increasing its point-mutation rate. They will use these mutants in their study of
mutation supply. -

Aryan Agarwal
HIGH SCHOOL STUDENT
Aryan is a high school student with us this summer. He is working with Shiva, using previous whole-genome sequence data from experimentally evolved S. aureus lines to derive a “heterogeneity
score,ˮ and assessing whether this metric
correlates with particular drug trade-offs.